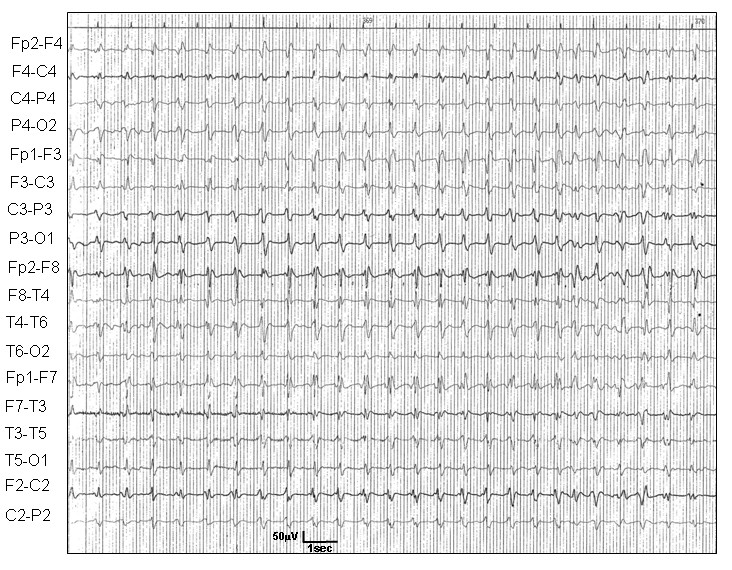
Brain-computer interaction is a rapidly growing technology that helps people with disabilities such as paralytic patients and provides solutions to those patients with neurological disabilities. The enthusiasm in this field has increased dramatically every day. The brain-computer interface (BCI) system provides users with a complete communication system between their bodies and external devices. It's a system in two ways. The imaginary motor process is a mental activity without any real movement of the body. Here, we will try to achieve a simple goal, given a moto imagery EEG signal, we will try to predict the class using machine learning.
load('SubA_5chan_3LRF.mat')% For DWTSubA_5C_LRF_c1 = zeros(270,1024);SubA_5C_LRF_c2 = zeros(270,1024);SubA_5C_LRF_c3 = zeros(270,1024);SubA_5C_LRF_c4 = zeros(270,1024);SubA_5C_LRF_c5 = zeros(270,1024);% 270 samples, 5 channels, 1024 data points% 1 response% 1024 = 256 * 4 = sampling rate * secfor smp = 1:270SubA_5C_LRF_c1(smp,:) = EEGDATA(1,:,smp);endfor smp = 1:270SubA_5C_LRF_c2(smp,:) = EEGDATA(2,:,smp);endfor smp = 1:270SubA_5C_LRF_c3(smp,:) = EEGDATA(3,:,smp);endfor smp = 1:270SubA_5C_LRF_c4(smp,:) = EEGDATA(4,:,smp);endfor smp = 1:270SubA_5C_LRF_c5(smp,:) = EEGDATA(5,:,smp);end
DWT_SubA_Channel5_coif2_3 = zeros(5,1024,270);for channel = 1:5for sample = 1:270DWT_SubA_Channel5_coif2_3(channel, :,...sample) = wden(EEGDATA(channel, :,...sample),'sqtwolog','s','mln',3,'coif2');endend
function feature_vec = feature_set_gen(mat_y)dwt_sig = mat_y;num_feature = 10;[~, data_pts, num_trial] = size(dwt_sig);dwt_sig = reshape(dwt_sig,[data_pts, num_trial]);dwt_sig = dwt_sig';feature_vec = zeros(num_trial,num_feature); % + 1 for response if usedfor nt = 1:num_trial% AAC (Avergae amplitude change)aac = 0;for i = 1:data_pts-1aac = aac + abs(dwt_sig(nt,i+1) - dwt_sig(nt,i));endaac = aac/data_pts;feature_vec(nt,1) = aac;% DASDV (Difference absolute standard deviation value)dasdv = 0;for i = 1:data_pts-1dasdv = dasdv + (dwt_sig(nt,i+1) - dwt_sig(nt,i))^2;enddasdv = dasdv/(data_pts-1);dasdv = sqrt(dasdv);feature_vec(nt,2) = dasdv;% Integrated EMG (IEMG)iemg = sum(abs(dwt_sig(nt,:)));feature_vec(nt,3) = iemg;% Mean absolute value (MAV)mav = iemg/data_pts;feature_vec(nt,4) = mav;% Modified mean absolute value Type 2(MMAV2)mmav2 = 0;for i = 1:data_ptstmp = abs(dwt_sig(nt,i));if i < 0.12*data_ptstmp = (tmp * 4 * i)/data_pts;mmav2 = mmav2 + tmp;endif i > 0.85*data_ptstmp = (tmp * 4 * (i-data_pts))/data_pts;mmav2 = mmav2 + tmp;endif i >= 0.12*data_pts && i<= 0.85*data_ptsmmav2 = mmav2 + tmp;endendmmav2 = mmav2/data_pts;feature_vec(nt,5) = mmav2;% Myopulse percentage rate (MYOP)myop_thres = 0.25;myop = (sum(dwt_sig(nt,:)>myop_thres))/data_pts;feature_vec(nt,6) = myop;% Root mean square (RMS)rms = sqrt(sum(dwt_sig(nt,:).^2)/data_pts);feature_vec(nt,7) = rms;% Slope sign change (SSC)ssc_thres = 0.08;ssc = 0;for i = 2:data_pts-1tmp1 = (dwt_sig(nt,i) - dwt_sig(nt,i-1))*(dwt_sig(nt,i) - dwt_sig(nt,i+1));if tmp1 >= ssc_thresssc = ssc + 1;endendfeature_vec(nt,8) = ssc;% Second spectral moment SM2sm2 = 0;rng default;[psd, fbin] = pwelch(dwt_sig(nt,:));bin_len = length(psd);for j = 1:bin_lensm2 = sm2 + psd(j)*(fbin(j)^2);endfeature_vec(nt,8) = sm2;% Log detector (LOG)logd = exp(sum(log(abs(dwt_sig(nt,:))))/data_pts);feature_vec(nt,9) = logd;% Waveform length (WL)wl = 0;for i = 1:data_pts-1wl = wl + abs(dwt_sig(nt,i+1) - dwt_sig(nt,i));endfeature_vec(nt,10) = wl;% feature_vec(nt,11) = resp;endendch1_dwt = DWT_SubA_Channel5_coif2_3(1,:,:);ch2_dwt = DWT_SubA_Channel5_coif2_3(2,:,:);ch3_dwt = DWT_SubA_Channel5_coif2_3(3,:,:);ch4_dwt = DWT_SubA_Channel5_coif2_3(4,:,:);ch5_dwt = DWT_SubA_Channel5_coif2_3(5,:,:);num_smp = 270;num_feature = 10;feature_c1 = feature_set_gen(ch1_dwt); % num_smp*featurefeature_c2 = feature_set_gen(ch2_dwt);feature_c3 = feature_set_gen(ch3_dwt);feature_c4 = feature_set_gen(ch4_dwt);feature_c5 = feature_set_gen(ch5_dwt);features_response_A = [feature_c1, feature_c2, feature_c3, ...feature_c4, feature_c5, LABELS];
function [trainedClassifier, validationAccuracy] = trainClassifier(datasetTable)% Convert input to tabledatasetTable = table(datasetTable);datasetTable.Properties.VariableNames = {'column'};% Split matrices in the input table into vectorsdatasetTable.column_1 = datasetTable.column(:,1);datasetTable.column_2 = datasetTable.column(:,2);datasetTable.column_3 = datasetTable.column(:,3);datasetTable.column_4 = datasetTable.column(:,4);datasetTable.column_5 = datasetTable.column(:,5);datasetTable.column_6 = datasetTable.column(:,6);datasetTable.column_7 = datasetTable.column(:,7);datasetTable.column_8 = datasetTable.column(:,8);datasetTable.column_9 = datasetTable.column(:,9);datasetTable.column_10 = datasetTable.column(:,10);datasetTable.column_11 = datasetTable.column(:,11);datasetTable.column_12 = datasetTable.column(:,12);datasetTable.column_13 = datasetTable.column(:,13);datasetTable.column_14 = datasetTable.column(:,14);datasetTable.column_15 = datasetTable.column(:,15);datasetTable.column_16 = datasetTable.column(:,16);datasetTable.column_17 = datasetTable.column(:,17);datasetTable.column_18 = datasetTable.column(:,18);datasetTable.column_19 = datasetTable.column(:,19);datasetTable.column_20 = datasetTable.column(:,20);datasetTable.column_21 = datasetTable.column(:,21);datasetTable.column_22 = datasetTable.column(:,22);datasetTable.column_23 = datasetTable.column(:,23);datasetTable.column_24 = datasetTable.column(:,24);datasetTable.column_25 = datasetTable.column(:,25);datasetTable.column_26 = datasetTable.column(:,26);datasetTable.column_27 = datasetTable.column(:,27);datasetTable.column_28 = datasetTable.column(:,28);datasetTable.column_29 = datasetTable.column(:,29);datasetTable.column_30 = datasetTable.column(:,30);datasetTable.column_31 = datasetTable.column(:,31);datasetTable.column_32 = datasetTable.column(:,32);datasetTable.column_33 = datasetTable.column(:,33);datasetTable.column_34 = datasetTable.column(:,34);datasetTable.column_35 = datasetTable.column(:,35);datasetTable.column_36 = datasetTable.column(:,36);datasetTable.column_37 = datasetTable.column(:,37);datasetTable.column_38 = datasetTable.column(:,38);datasetTable.column_39 = datasetTable.column(:,39);datasetTable.column_40 = datasetTable.column(:,40);datasetTable.column_41 = datasetTable.column(:,41);datasetTable.column_42 = datasetTable.column(:,42);datasetTable.column_43 = datasetTable.column(:,43);datasetTable.column_44 = datasetTable.column(:,44);datasetTable.column_45 = datasetTable.column(:,45);datasetTable.column_46 = datasetTable.column(:,46);datasetTable.column_47 = datasetTable.column(:,47);datasetTable.column_48 = datasetTable.column(:,48);datasetTable.column_49 = datasetTable.column(:,49);datasetTable.column_50 = datasetTable.column(:,50);datasetTable.column_51 = datasetTable.column(:,51);datasetTable.column = [];% Extract predictors and responsepredictorNames = {'column_1', 'column_2', 'column_3', 'column_4', 'column_5', 'column_6', 'column_7', 'column_8', 'column_9', 'column_10', 'column_11', 'column_12', 'column_13', 'column_14', 'column_15', 'column_16', 'column_17', 'column_18', 'column_19', 'column_20', 'column_21', 'column_22', 'column_23', 'column_24', 'column_25', 'column_26', 'column_27', 'column_28', 'column_29', 'column_30', 'column_31', 'column_32', 'column_33', 'column_34', 'column_35', 'column_36', 'column_37', 'column_38', 'column_39', 'column_40', 'column_41', 'column_42', 'column_43', 'column_44', 'column_45', 'column_46', 'column_47', 'column_48', 'column_49', 'column_50'};predictors = datasetTable(:,predictorNames);predictors = table2array(varfun(@double, predictors));response = datasetTable.column_51;% Train a classifiertemplate = templateSVM('KernelFunction', 'polynomial', 'PolynomialOrder', 3, 'KernelScale', 'auto', 'BoxConstraint', 1, 'Standardize', 1);trainedClassifier = fitcecoc(predictors, response, 'Learners', template, 'Coding', 'onevsone', 'PredictorNames', {'column_1' 'column_2' 'column_3' 'column_4' 'column_5' 'column_6' 'column_7' 'column_8' 'column_9' 'column_10' 'column_11' 'column_12' 'column_13' 'column_14' 'column_15' 'column_16' 'column_17' 'column_18' 'column_19' 'column_20' 'column_21' 'column_22' 'column_23' 'column_24' 'column_25' 'column_26' 'column_27' 'column_28' 'column_29' 'column_30' 'column_31' 'column_32' 'column_33' 'column_34' 'column_35' 'column_36' 'column_37' 'column_38' 'column_39' 'column_40' 'column_41' 'column_42' 'column_43' 'column_44' 'column_45' 'column_46' 'column_47' 'column_48' 'column_49' 'column_50'}, 'ResponseName', 'column_51', 'ClassNames', [1 2 3]);% Set up holdout validationcvp = cvpartition(response, 'Holdout', 0.25);trainingPredictors = predictors(cvp.training,:);trainingResponse = response(cvp.training,:);% Train a classifiertemplate = templateSVM('KernelFunction', 'polynomial', 'PolynomialOrder', 3, 'KernelScale', 'auto', 'BoxConstraint', 1, 'Standardize', 1);validationModel = fitcecoc(trainingPredictors, trainingResponse, 'Learners', template, 'Coding', 'onevsone', 'PredictorNames', {'column_1' 'column_2' 'column_3' 'column_4' 'column_5' 'column_6' 'column_7' 'column_8' 'column_9' 'column_10' 'column_11' 'column_12' 'column_13' 'column_14' 'column_15' 'column_16' 'column_17' 'column_18' 'column_19' 'column_20' 'column_21' 'column_22' 'column_23' 'column_24' 'column_25' 'column_26' 'column_27' 'column_28' 'column_29' 'column_30' 'column_31' 'column_32' 'column_33' 'column_34' 'column_35' 'column_36' 'column_37' 'column_38' 'column_39' 'column_40' 'column_41' 'column_42' 'column_43' 'column_44' 'column_45' 'column_46' 'column_47' 'column_48' 'column_49' 'column_50'}, 'ResponseName', 'column_51', 'ClassNames', [1 2 3]);% Compute validation accuracyvalidationPredictors = predictors(cvp.test,:);validationResponse = response(cvp.test,:);validationAccuracy = 1 - loss(validationModel, validationPredictors, validationResponse, 'LossFun', 'ClassifError');