In simple words, ECG represents the electrical activity of the heart and in this article, we will try to separate normal ECG patterns and abnormal ECG with the help of machine learning. We have to extract some features so that machine learning models can learn from those features. We will be using spectral moment 2, LOG, wavelength length, autocorrelation, binned entropy, sample entropy, time-reversal asymmetry statistic, and variance. We will be using multiple machine learning models to benchmark our results. Let's using SVM, KNN, Decision Tree, Random Forest, AdaBoost from the scikit-learn library. We can compare the accuracy of all the models. We will be using a 5 fold cross-validation.
In simple words, ECG represents the electrical activity of heart and in this article, we will try to separate normal ECG patterns and abnormal ECG with the help of machine learning.
from scipy import io, signalimport matplotlib.pyplot as pltimport dtcwtimport numpy as npimport itertoolsimport pywt
test_path = 'MLII/reformatted_dataset/normal/100m (0)_nsr.mat'ta = io.loadmat(test_path)print(ta['val'])[[953 951 949 ... 940 943 944]]print(ta){'val': array([[953, 951, 949, ..., 940, 943, 944]], dtype=int16)}ta = ta['val']print(type(ta))ta.shape
(1, 3600)print(ta)[[953 951 949 ... 940 943 944]]import numpy as npta = np.array(ta)ta = np.reshape(ta, (3600,))
import matplotlib.pyplot as pltplt.plot(ta)plt.show()
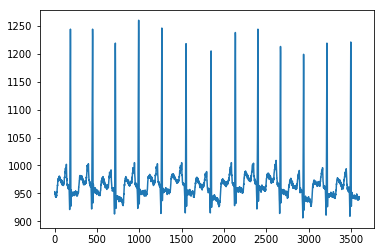
def plot_ecg(path, tit):ta = io.loadmat(path)ta = ta['val']ta = np.array(ta)ta = np.reshape(ta, (ta.shape[1],))plt.plot(ta)plt.title(tit)plt.show()
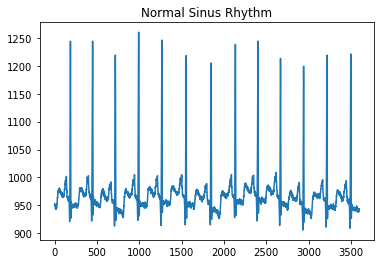
def get_ecg(path):ta = io.loadmat(path)ta = ta['val']ta = np.array(ta)ta = np.reshape(ta, (ta.shape[1],))return taplot_ecg('MLII/reformatted_dataset/normal/100m (0)_nsr.mat', 'Normal Sinus Rhythm')
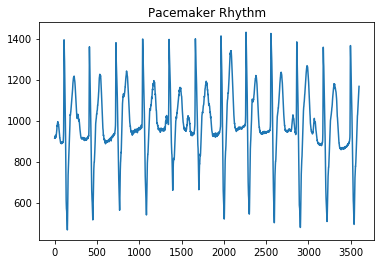
plot_ecg('MLII/reformatted_dataset/normal/107m (5)_pr.mat', 'Pacemaker Rhythm')
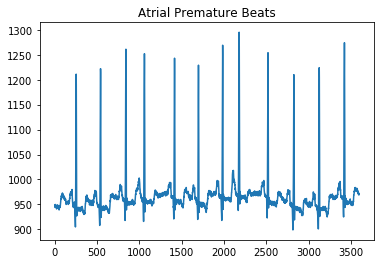
plot_ecg('MLII/reformatted_dataset/arythmia/100m (0)_apb.mat', 'Atrial Premature Beats')
from pywt import wavedeccoeffs = wavedec(x, 'db4', level=2)cA2, cD2, cD1 = coeffsplt.plot(cA2)plt.show()plt.plot(cD2)plt.show()plt.plot(cD1)plt.show(
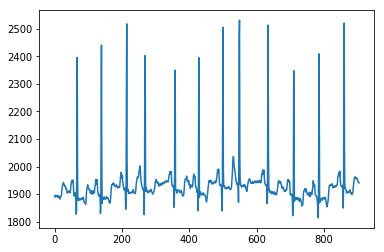
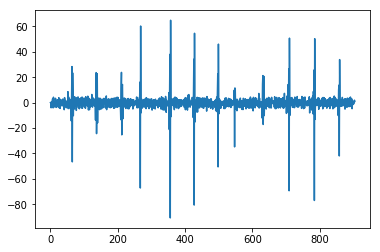
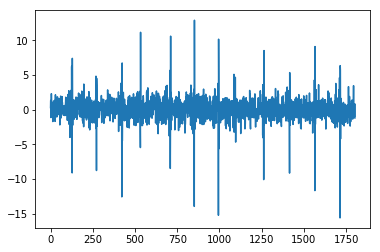
import pandas as pdfrom numpy.linalg import LinAlgErrorfrom statsmodels.tsa.stattools import adfuller#1def AE(x): # Absolute Energyx = np.asarray(x)return sum(x * x)#2def SM2(y):#t1 = time.time()f, Pxx_den = signal.welch(y)sm2 = 0n = len(f)for i in range(0,n):sm2 += Pxx_den[i]*(f[i]**2)#t2 = time.time()#print('time: ', t2-t2)return sm2#3def LOG(y):n = len(y)return np.exp(np.sum(np.log(np.abs(y)))/n)#4def WL(x): # WL in primary manuscriptreturn np.sum(abs(np.diff(x)))#6def AC(x, lag=5): # autocorrelation""""""# This is important: If a series is passed, the product below is calculated# based on the index, which corresponds to squaring the series.if type(x) is pd.Series:x = x.valuesif len(x) < lag:return np.nan# Slice the relevant subseries based on the lagy1 = x[:(len(x)-lag)]y2 = x[lag:]# Subtract the mean of the whole series xx_mean = np.mean(x)# The result is sometimes referred to as "covariation"sum_product = np.sum((y1-x_mean)*(y2-x_mean))# Return the normalized unbiased covariancereturn sum_product / ((len(x) - lag) * np.var(x))#7def BE(x, max_bins=30): # binned entropyhist, bin_edges = np.histogram(x, bins=max_bins)probs = hist / len(x)return - np.sum(p * np.math.log(p) for p in probs if p != 0)#15def SE(x): # sample entropy""""""x = np.array(x)sample_length = 1 # number of sequential points of the time seriestolerance = 0.2 * np.std(x) # 0.2 is a common value for r - why?n = len(x)prev = np.zeros(n)curr = np.zeros(n)A = np.zeros((1, 1)) # number of matches for m = [1,...,template_length - 1]B = np.zeros((1, 1)) # number of matches for m = [1,...,template_length]for i in range(n - 1):nj = n - i - 1ts1 = x[i]for jj in range(nj):j = jj + i + 1if abs(x[j] - ts1) < tolerance: # distance between two vectorscurr[jj] = prev[jj] + 1temp_ts_length = min(sample_length, curr[jj])for m in range(int(temp_ts_length)):A[m] += 1if j < n - 1:B[m] += 1else:curr[jj] = 0for j in range(nj):prev[j] = curr[j]N = n * (n - 1) / 2B = np.vstack(([N], B[0]))# sample entropy = -1 * (log (A/B))similarity_ratio = A / Bse = -1 * np.log(similarity_ratio)se = np.reshape(se, -1)return se[0]#16def TRAS(x, lag=5):# time reversal asymmetry statistic"""| [1] Fulcher, B.D., Jones, N.S. (2014).| Highly comparative feature-based time-series classification.| Knowledge and Data Engineering, IEEE Transactions on 26, 3026–3037."""n = len(x)x = np.asarray(x)if 2 * lag >= n:return 0else:return np.mean((np.roll(x, 2 * -lag) * np.roll(x, 2 * -lag) * np.roll(x, -lag) -np.roll(x, -lag) * x * x)[0:(n - 2 * lag)])#17def VAR(x): # variancereturn var(x)
from sklearn.preprocessing import StandardScalerscaler = StandardScaler()scaler.fit(fx)print(scaler.mean_)X_all = scaler.transform(fx)print(np.mean(X_all))print(np.std(X_all))
from sklearn.model_selection import cross_val_scorefrom sklearn import svmclf = svm.SVC(kernel='linear', C=1)scores = cross_val_score(clf, X_all, y, cv=5)print('Accuracy: ', scores.mean(), scores.std() * 2)Accuracy: 0.7272051167633496 0.1763962471741112from sklearn.neighbors import KNeighborsClassifierknn = KNeighborsClassifier(n_neighbors=100)scores = cross_val_score(knn, X_all, y, cv=5)print('Accuracy: ', scores.mean(), scores.std() * 2)Accuracy: 0.7980960880559274 0.007141157066570031from sklearn import treeclf = tree.DecisionTreeClassifier()scores = cross_val_score(clf, X_all, y, cv=5)print('Accuracy: ', scores.mean(), scores.std() * 2)Accuracy: 0.7046854082998661 0.17689499484705223from sklearn.ensemble import RandomForestClassifierclf = RandomForestClassifier(n_estimators=100, max_depth=None,min_samples_split=4, random_state=0)scores = cross_val_score(clf, X_all, y, cv=5)print('Accuracy: ', scores.mean(), scores.std() * 2)Accuracy: 0.734374535177748 0.2567492330745884from sklearn.ensemble import AdaBoostClassifierclf = AdaBoostClassifier(n_estimators=10)scores = cross_val_score(clf, X_all, y, cv=5)print('Accuracy: ', scores.mean(), scores.std() * 2)